research
My research explores how individual heterogeneity and spatial structure generate rare but consequential events that disproportionately shape ecological and epidemiological dynamics. I’m driven by understanding nature through numbers—finding creative solutions when faced with data limitations, uncertainty, or complex systems that resist traditional approaches.
I’m model-agnostic: I select the best tool for the question rather than adhering to a single framework. My toolkit spans extreme value statistics, Bayesian hierarchical models, spatially-explicit simulations, machine learning, and decision science. I’m committed to open science, and when I have a say, my work is publicly available with annotated code on GitHub.
Research Themes
Individual Heterogeneity & Extreme Dispersal Events
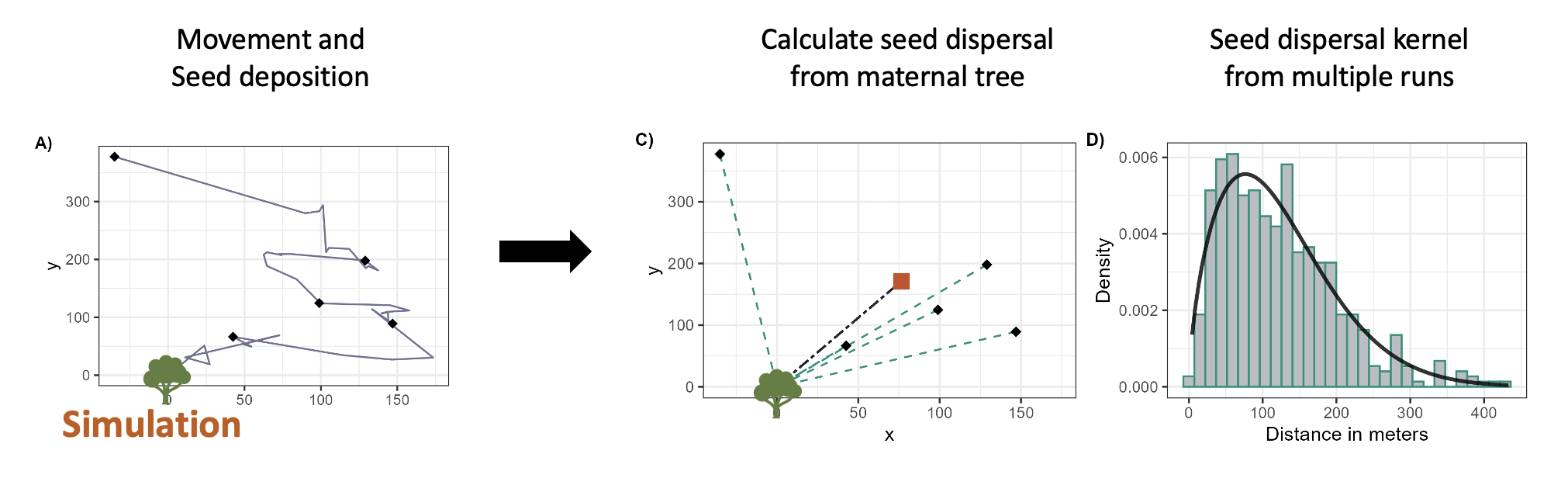
Rare individuals drive disproportionate ecological outcomes, whether through long-distance seed dispersal or superspreading events in disease transmission. My dissertation developed frameworks using extreme value theory (EVT) to characterize individual heterogeneity in animal movement, quantifying not average behavior but the tail dynamics that generate rare, high-impact events.
This theoretical foundation transfers directly to multihost disease systems: superspreaders exist in the tail of contact or movement distributions, and their identification requires methods designed for extremes rather than central tendencies. I’m extending this work by integrating step selection analysis with mechanistic transmission models to ask: When does individual heterogeneity amplify versus dampen outbreak risk across host species? How do landscape features interact with movement heterogeneity in bridge host populations? Can we develop movement-based surveillance strategies that predict spillover before emergence?
I’m presenting this work at UF’s Animal Behavior and Infectious Disease Symposium (March 2026), focusing on EVT applications to detecting cryptic superspreading dynamics in multihost systems, a methodological contribution that connects movement ecology, network epidemiology, and surveillance optimization.
Hybrid Inference for Resource-Limited Surveillance

Mechanistic epidemiological models need fine-scale data. Real-world surveillance produces yearly case totals, aggregated counts, or opportunistic observations. I develop hybrid frameworks integrating statistical inference, data augmentation, and mechanistic simulations to bridge this gap.
Demonstrated systems: - Anthrax in Vietnam: Semi-synthetic data augmentation fitting mechanistic models to yearly totals (One Health, 2025) - White-nose syndrome in Montana: Structured decision-making under deep uncertainty through stakeholder engagement (in review) - SARS-CoV-2 in deer: Spillover risk and transmission dynamics between wild and farmed deer (PLOS Computational Bio, 2024)
Current work: Expanding to multi-host, multi-pathogen systems at biological invasion frontiers. Through my Global Fellows Award, I’m establishing research collaborations in southern Chile examining invasive species as potential pathogen bridge hosts at the livestock-wildlife interface and formalizing this partnership during an extended research visit in May 2026. This work positions me to develop computational frameworks for detecting bridge host roles using sparse surveillance data, with future proposals planned to scale these approaches across hemispheric invasion contexts.
Landscape Geometry as Active Driver of Transmission
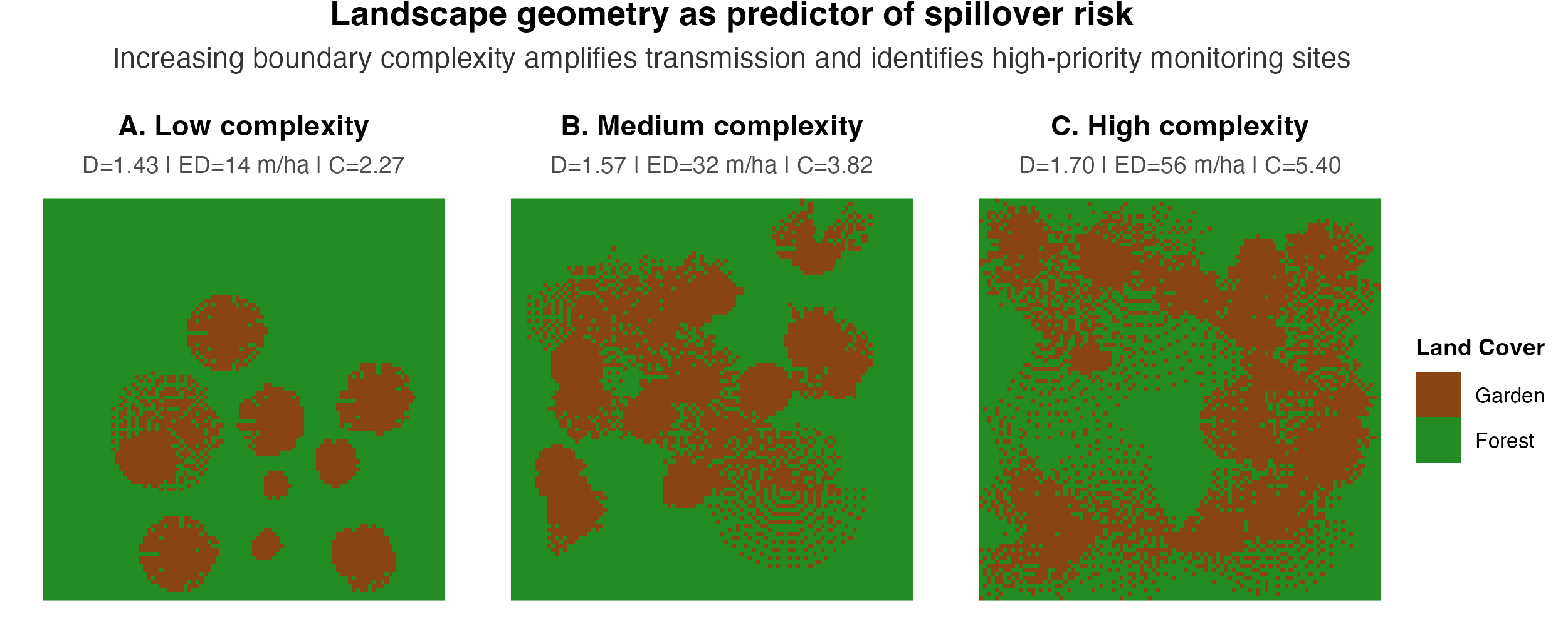
Classical disease ecology treats landscape as passive backdrop. I’m developing frameworks positioning landscape geometry (fractal dimension, patch configuration, edge density) as an active transmission driver in mechanistic epidemiological models.
My approach integrates geometric landscape features directly into spatially-explicit transmission models, creating bidirectional inference: simulate forward (landscape geometry → disease patterns) and work backward through inverse modeling (observed disease dynamics → geometric signatures). This reframes landscape geometry as an inferential target rather than requiring exhaustive field surveys—essential for systems where remote sensing data is abundant but disease surveillance remains sparse.
In collaboration with the Florida Museum of Natural History, I’m applying these frameworks to Amazonian systems using satellite-derived landscape metrics, with future applications to vector-borne diseases in fragmented forests, zoonotic spillover at urban-wildland interfaces, and waterborne diseases in canal systems.
Metacommunity Theory Meets Disease Ecology
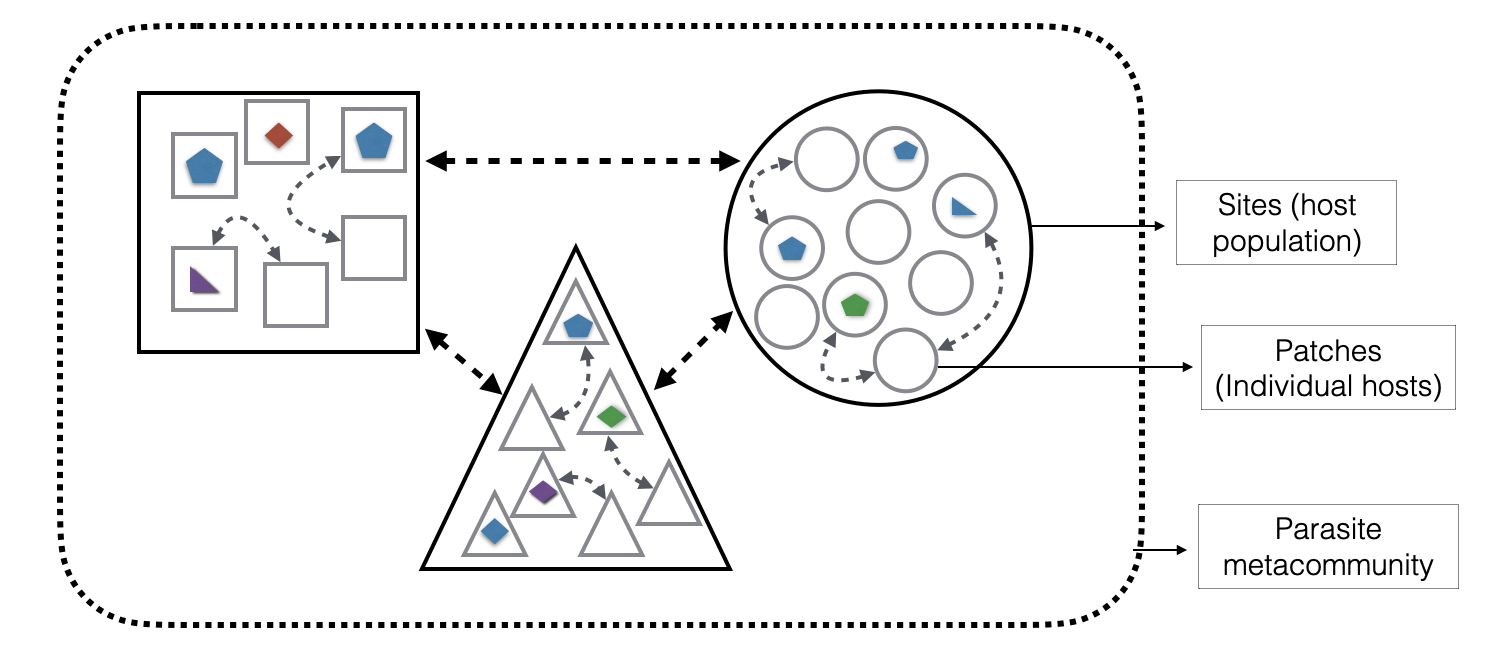
For several years I’ve explored parallels between metacommunity ecology and disease ecology: treating pathogens as ecological communities subject to assembly rules across host metacommunities.
My work explores how species-specific dispersal variation affects community assembly and whether JSDMs can detect these processes, building on our published work for metacommunity theory. I co-teach UF’s Advanced Ecology: Theory and Computational Methods course with Dr. Mathew Leibold, where we teach JSDM approaches for complex ecological systems and explore extensions to their applications and links to macroecology.
Disease ecology application: This framework has the potential to enable surveillance optimization for multi-host zoonotic systems. By modeling pathogens probabilistically across host communities, we can identify which host species and populations maximize information about pathogen community structure when comprehensive monitoring is prohibitively expensive, critical for systems where dozens of pathogen strains circulate across wildlife, livestock, and human populations.
Methodological Journey
My research program emerged from a methodological journey spanning probability theory, hierarchical modeling, and mechanistic simulation. During my PhD (UF Biology/Zoology), I developed expertise in extreme value statistics and simulation techniques. My postdoctoral work at USGS Patuxent and UF’s Emerging Pathogens Institute expanded this to Bayesian hierarchical models, occupancy modeling, and mechanistic disease models integrated with machine learning.
As Computational Literacy Librarian at UF’s Academic Research & Consulting Services, I now study how computational methods themselves transfer across biological systems, what I call methodology science. This positioning allows me to establish international research collaborations that build computational capacity across entire research communities, not just individual projects.
Computational Capacity Building in Latin America
My 2026 Global Fellows Award supports establishing computational disease ecology training networks across Latin America, with workshops planned for Chile (May 2026) and Ecuador (July 2026). These workshops are designed for variable computational access and resource-limited contexts, with all materials developed as open educational resources available to UF students and international partners.
I’m currently mentoring two undergraduate researchers developing these frameworks: one creating an OER module on SIR models and disease ecology, another building reproducible research templates in R and GitHub in collaboration with UF’s Environmental and Global Health Department. This work positions UF as a hemispheric hub for computational approaches to One Health challenges.
Open Science Commitment
Most of my work is publicly available with annotated code on GitHub. I’ve promoted reproducibility in ecology for years and develop materials emphasizing transparent, reproducible workflows. Exceptions occur when data are regulated by government agencies or I’m not primary owner, but transparency is my default.